Neuroscience¶
Neuronal energetics¶
On of the goals of my first postdoc project was to understand how energy consumption influences neuronal activity and vice-versa. To that end, I designed new dynamical models to model the interactions between membrane potential, calcium, and energy production/consumption and analyze how energy regulation affects the overall ability of the network to process and transfer information, as well as adapt to changes in the input patterns.
Contrary to previous work on energetic constraints (which often focused on global energetic cost, or on synaptic transmission), my work focuses on the local dynamics in the region of the soma and axon initial segment.
Because many brain networks do not function in conditions which would place the neurons under signigicant energetic stress, my work is mainly focused on two specific cases:
epileptic seizures and epileptiform bursts of activity in neuronal cultures,
Parkinsonian networks and the implication of the energy crisis associated to this disease.
Link to the lab's webpage.
Modeling energy¶
Two initial models are available on three simulation platforms (NEST, BRIAN, and NEURON). Source code can be found here and the models are discussed in details in a paper in PLoS CB, available on BioRxiv (DOI: 10.1371/journal.pcbi.1008503).
PhD project¶
My goal was to understand what is required for neuronal populations to be able to process information, then to be trained (for instance to reproduce perceptron-like capabilities).
Before turning to signal processing and learning, the NeuroPhysics team, where I was working, focused on the basic behavior of neuronal cultures. Indeed, the idea was to understand what drives collective behaviors of freely evolving cultures, so that we can obtain insights on how one can either exploit this intrinsic dynamics or prevent it from occuring.
To that end, I used tools from dynamical systems and neuronal simulations to reproduce the experimental observation of our collaborators and other experimental teams around the world (see details on the Neuronal activity section).
Structuring the network seems a promising way to control the overall activity of a culture. Moreover, it is experimentally feasible through microfabrication, which is why we are considering it of prime importance. In order to predict the activity of such patterned culture and to compare our simulations to experimental observations, I developed a parallel simulator to model the growth of neurons inside a culture (see Growing neurons section).
Using realistic network structures on which I will simulate neuronal activity, I aim at predicting structures that would allow for efficient separation of different input signals and learning.
Update the manuscript is now online! You can find it here or download it directly by clicking on the image below.

Neuronal activity¶
In order to fulfill this goal of making neuronal cultures process information, we must first understand the origin of the behavior observed in isolated neuronal cultures. Once unconstrained cultures are understood, we can then determine how their intrinsic behavior can be used or modulated in order to process afferent inputs.
Synchronization in neuronal cultures¶
A common type of activity observed in neuronal cultures is a synchronized state were the neurons fire in phase. This activity is mostly observed under the form of synchronized bursts, where the neurons fire several spikes on a short time window, then become quiet over a long period of time before they start firing again.
Simple model: neurons as adaptive oscillators¶
Several studies have shown the presence of self-ocillating neurons, called pacemaker neurons, in neuronal networks. Considering a population of such neurons, I demonstrated in a 2018 paper how spike-driven adaptation can shape an oscillating activity into well-defined and synchronous network bursts.
What drives the initiation and termination of such bursts?¶
Using dynamical systems and phase-plane analysis, I demonstrated that, in an adaptive integrate-and-fire model, the termination of a burst is determined by the hyperpolarizing currents following from the series spikes emitted by a neuron during the burst. In this model, the termination is the source of the neuronal synchronisation, while the initiation of the subsequent burst caused by persistent currents, such as I_{Na,p} or I_M, which drive the progressive depolarization of the membrane and makes the first neurons fire. These first neurons trigger (advance) the firing of the rest of the network.
Propagation of the activity to the whole network¶
How a whole neuronal population is recruited into a network burst from the first few firing neurons has been extensively discussed in the litterature, notably in the framework of percolation theory.
In a paper published in Physica A, we demonstrated the relevance of this model in the presence of inhibitory neurons, showing that their presence was simply equivalent to making the network less excitable. In this paper, we also showed that dynamical integrate-and-fire neurons also followed this universal percolation behavior.
Hence, above a certain level of excitation, unstructured network will necessarily give rise to such network-wide bursting events.
See Chapter 3 in the my thesis and continued work in the thesis of Mallory Dazza and work by Anna Levina and Oleg Vinogradov for details.
Growing neurons¶
Based on our result on the influence of adaptation and pacemaker neurons, the emergent bursting behavior should prove very difficult to repress in unstructured cultures if they are triggered by pacemaker neurons, that is unless a way of removing the pacemaker neurons can be found. To tackle this issue, I propose to use patterned cultures, where the connectivity would be structured in such a way that the excitation from the pacemaker neurons cannot propagate and recruit the whole network.
Spatial cultures¶
In order to take the relevant spatiotemporal correlations that are present in neuronal cultures into account, the networks that we study must contain the spatial information of the cell positions and connection lengths.
In homogeneous cultures, this can be performed rather effectively using generative models, and the NNGT library allows versatile network generation in spatial environments. This first step allowed us to study the detailed spatiotemporal dynamics of burst nucleation in homogeneous cultures.
However, such simple models become more and more inaccurate as the spatial complexity increases, especially in structured cultures, where obstacles can block the neurites' path, making the actual connection distance often much larger than the euclidean distance between the two neurons.
DeNSE: a new growth simulator¶
To properly take such complex spatial environments into account, the main requirement is the implementation of a parallel simulator, DeNSE, which can model the growth process of neurons in 2D cultures. Moreover, beyond spatial cultures, the goal of this simulator is also to provide biologically detailed mechanisms for neurite outgrowth so that it can help isolate relevant factors in the growing process.
As of July 2019, a pre-release of the simulator is available on GitHub (https://github.com/SENeC-Initiative/DeNSE) and its documentation is present on ReadTheDocs (https://dense.readthedocs.io/en/latest/).
Simple neurons¶
Examples of structures obtained through the DeNSE simulator. On the left is a multipolar neuron with limited arborization obtained through lateral branching. One the right is a purkinje-like cell displaying an exhuberant dendritic arbor which stems from a combination of initial splits and later interstitial branching events. Axons are marked in red, dendrites in blue, somas in black. Scale bars are 50 :math:`mu`m.
Space-embedded populations¶
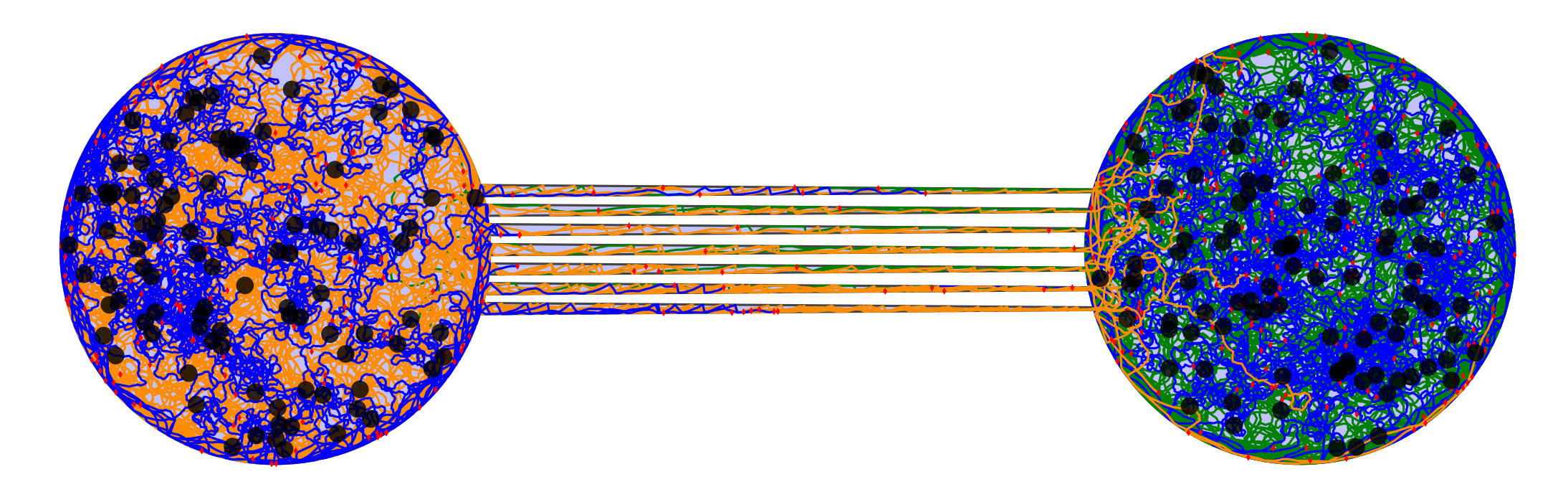
Two chambers with asymmetrical funnels formign an effective neuronal diode: axons from the left chamber can innervate the right chamber while the reverse is a lot less likely.
On the simulated cultures, dendrites are in blue, left axons in orange and right axons in green.
See DeNSE and associated abstract and poster PDF for more details. Some (older) discussions are also available in the manuscript of my PhD project, Chapter 4.